Permutation Weighting for Estimating Marginal Structural Model Parameters
A few weeks ago, I demonstrated how permutation weighting (Arbour, Dimmery, and Sondhi 2020) can be used to estimate causal parameters under interference. Today, I show how to use the same idea to estimate parameters in a longitudinal marginal structural model (MSM) (Robins 1999; Robins, Hernán, and Brumback 2000). This post is mostly me proving to myself that it works when combining interference with longitudinal data in a MSM.
I’m not going to go in depth on the theory or purpose of MSMs here. But briefly, MSMs have traditionally been applied in a longitudinal data context. The analyst supposes a parametric relationship between average potential outcomes; for example, a simple MSM for a continuous potential outcome under an intervention from two previous timepoints is:
Under standard causal assumptions (such as no unnmeasured confounding), the s have a causal interpretation and can be estimated from the model using (stabilized) inverse probability weights taken over a subject’s history. The weights we’re trying to estimate are analogous to equation (2) in Arbour, Dimmery, and Sondhi (2020) with the difference that we need to maintain the clustered nature (within an individual) of treatments rather then permuting all the rows independently.
Simulation Study
I’m hiding the simulator functions, as the code is kludgy, similar to the 2020-01-19 post, and not the point, but you can find the source
.Rmdon github.
To make things interesting, the simulation simply extends the interference simulation by adding additional timepoints for each subject. The outcomes can depend on the unit’s exposure (and that of neighbors) from the current and previous timepoint, as well as the outcome from the previous timepoint. To get time-varying confounding, a unit’s exposure also depends on the previous exposure.
Generic Permutation Weighting Functions
#' Stack the observed data with the permuted data
#' @export
permute_and_stack <- function(dt, permuter){
dplyr::bind_rows(
dplyr::mutate(dt, C = 0),
dplyr::mutate(permuter(dt), C = 1)
)
}
#' Estimate the density ratio by modeling whether an observation is from
#' the permuted dataset or original dataset
.pw <- function(dt, rhs_formula, fitter, modify_C, predictor, fitter_args, permuter){
pdt <- permute_and_stack(dt, permuter)
pdt$C <- modify_C(pdt$C)
m <- do.call(
fitter,
args = c(list(formula = update(C ~ 1, rhs_formula), data = pdt), fitter_args))
w <- predictor(object = m, newdata = dt)
w/(1 - w)
}
#' Estimate permutation weights B times and average the results
#' @export
get_permutation_weights <- function(dt, B, rhs_formula, fitter, modify_C = identity,
predictor, fitter_args = list(),
permuter){
out <- replicate(
n = B,
expr = .pw(dt, rhs_formula = rhs_formula, fitter = fitter, modify_C = modify_C,
predictor = predictor, fitter_args = fitter_args,
permuter = permuter)
)
apply(out, 1, mean)
}
#' Create a permutation weighted estimator for the marginal structural model
#' @export
make_pw_estimator <- function(fitter, rhs_formula, B,
modify_C = identity,
predictor = function(object, newdata) {
predict(object = object, newdata = newdata, type = "response")
},
fitter_args = list(),
permuter){
function(data){
w <- get_permutation_weights(
dt = data, B = B, rhs_formula = rhs_formula, fitter = fitter,
modify_C = modify_C, predictor = predictor, fitter_args = fitter_args,
permuter = permuter
)
w
}
}Specific PW functions
These functions are specific for analyzing the data simulated for this post. Note that the permuting function permutes entire vectors of treatment. Also the binary classifier used in the permutation weighting estimator uses a GEE model.
# Half ass permutation attempt with several poor practices
# * number of timepoints is hardcoded!
# * number of timepoints is same for all subjects
# * object `edges` will need to be found in an another environment (i.e. Global)
permuterf <- function(dt){
dt <- dplyr::arrange(dt, id, t)
rl <- rle(dt$id)
permutation <- rep((sample(rl$values, replace = FALSE) - 1L) * 4L, each = 4L) + 1:4
dt %>%
dplyr::mutate(
A = A[permutation]
) %>%
dplyr::group_by(t) %>%
dplyr::mutate(
# Number of treated neighbors
A_n = as.numeric(edges %*% A),
# Proportion of neighbors treated
fA = A_n/m_i,
id = id + max(id)
) %>%
dplyr::ungroup() %>%
dplyr::group_by(id) %>%
dplyr::mutate(
Al = dplyr::lag(A),
fAl = dplyr::lag(fA)
) %>%
dplyr::ungroup()
}
gee_fitter <- function(formula, data, ...){
data <- dplyr::filter(data, t > 0)
geepack::geeglm(formula = formula, data = data, id = id, family = binomial)
}
pdct <- function(object, newdata) {
predict(object = object, newdata = dplyr::filter(newdata, t > 0), type = "response")
}Here’s the estimator I’ll use:
pw_geeimator <- make_pw_estimator(
gee_fitter,
B = 5,
rhs_formula = ~ ((A + fA + Al + fAl):(factor(t) + Z1_abs*Z2 + Z3 + Yl) +
(factor(t) + Z1_abs*Z2 + Z3 + Yl)),
permuter = permuterf,
predictor = pdct)# Fix an adjacency matrix for use across simulations
# Set sample size to 5000
edges <- gen_edges(n = 5000, 5)
sim_data <- function(){
edges %>%
gen_data_0(
gamma = c(-2, 0.2, 0, 0.2, 0.2),
beta = c(2, 0, 0, -1.5, 2, -3, -3)
) %>%
purrr::reduce(
.x = 1:3, # 3 timepoints after time 0
.f = ~ gen_data_t(.x, .y,
gamma = c(-2, 3, 0, 0.2, 0, 0.2, 0.2),
beta = c(2, 0, 0, 0, -0.5, 0, -1.5, 2, -3, -3)),
.init = .
) %>%
purrr::pluck("data") %>%
dplyr::arrange(id, t)
}
do_sim <- function(){
dt <- sim_data()
w <- pw_geeimator(dt)
dt <- dt %>% dplyr::filter(t > 0) %>% dplyr::mutate(w = w)
res_w <- geepack::geeglm(
Y ~ -1 + factor(t) + A + Al + fA + fAl, data = dt, id = id, weights = dt$w
)
res_naive <- geepack::geeglm(
Y ~ -1 + factor(t) + A + Al + fA + fAl, data = dt, id = id
)
bind_rows(
broom::tidy(res_naive) %>% dplyr::mutate(estimator = "naive"),
broom::tidy(res_w) %>% dplyr::mutate(estimator = "pw_weighted")
)
}In the sim_data function, I set the parameters so that the only non-null effect is the exposure of neighbors from the previous timepoint.
Results
The plot below shows the bias from 100 simulations for the 4 causal parameters from a naive (unweighted) GEE model and the permutation weighted version.
library(dplyr)
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
library(geepack)
library(ggplot2)
res <-
purrr::map_dfr(1:100, ~ do_sim(), .id = "simid") %>%
filter(grepl("A", term)) %>%
group_by(simid) %>%
mutate(
bias = estimate - c(0, 0, 0, -0.5)
)
ggplot(
data = res,
aes(x = estimator, y = bias)
) +
geom_hline(yintercept = 0) +
geom_jitter() +
facet_grid(~ term)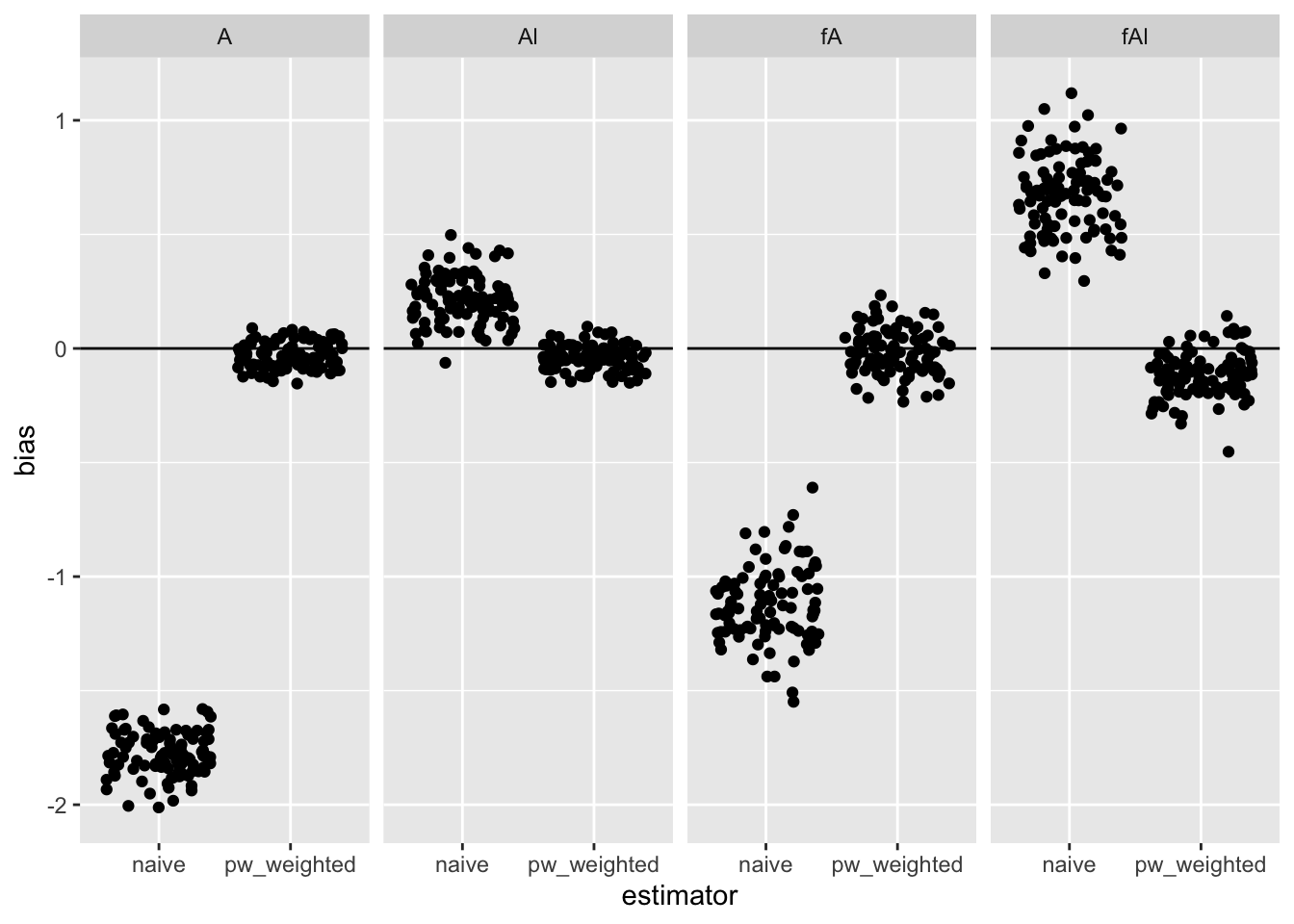
Summary
- Even after adding a time element to interference simulation from the other day, permutation weighting still works. I’m not surprised.
- The estimate for the non-null
fAlappears downwardly biased a bit. I’ll want to look into that further, but in general permutation weighting seems like a promising approach.